Tutorial on plant pollinator data
tutorial-bipartite.RmdEstimation with colSBM
We load a list of 15 plant-pollinator networks. They are binary undirected networks with different number of plant and pollinator species.
Networks benefiting from joint modelisation
First, we are going to model jointly the medan2002ld, medan2002rb networks, using the iid-colBiSBM model.
set.seed(1234, "L'Ecuyer-CMRG")
res_pp_iid <- estimate_colBiSBM(
netlist = dorebipartite[7L:8L], # A list of networks
colsbm_model = "iid", # The name of the model
net_id = names(dorebipartite)[7L:8L], # Name of the networks
nb_run = 2L, # Number of runs of the algorithm
global_opts = list(
verbosity = 1L,
plot_detail = 0L,
nb_cores = 2L,
backend = "parallel"
)
)
#>
#> Merging the 2 models
#> After merging the 2 model runs, the criteria are the following:
#>
#> vbound :
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] -725 -625 -620 -620 -Inf
#> [2,] -720 -614 -606 -Inf -Inf
#> [3,] -720 -607 -596 -Inf -Inf
#> [4,] -Inf -Inf -Inf -Inf -Inf
#> [5,] -Inf -Inf -Inf -Inf -Inf
#> [6,] -Inf -Inf -Inf -Inf -Inf
#>
#> ICL :
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] -729 -635 -642 -653 -Inf
#> [2,] -733 -651 -668 -Inf -Inf
#> [3,] -740 -686 -667 -Inf -Inf
#> [4,] -Inf -Inf -Inf -Inf -Inf
#> [5,] -Inf -Inf -Inf -Inf -Inf
#> [6,] -Inf -Inf -Inf -Inf -Inf
#>
#> BICL :
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] -729 -635 -636 -642 -Inf
#> [2,] -730 -634 -636 -Inf -Inf
#> [3,] -736 -638 -640 -Inf -Inf
#> [4,] -Inf -Inf -Inf -Inf -Inf
#> [5,] -Inf -Inf -Inf -Inf -Inf
#> [6,] -Inf -Inf -Inf -Inf -Inf
#>
#> Best fit at Q=( 2, 2 )
#>
#>
#>
#> ==== Best fits criterion for the 2 networks. Computed in 0.369 secs ====
#> Sep BiSBM total BICL: -640.6602
#> colBiSBM BICL: -633.8671
#> Joint modelisation preferred. With Q = ( 2, 2 ).
#>
#> ==== Full computation performed in 10.5 secs ====The output indicates that the collection benefits from a joint modelisation
Joint modelisation preferred
This is based on the BICL criterion.
We can look at how the variational bound and the model selection criteria evolve with the number of clusters. Here, the BICL criterion selects Q = 2, 2 blocks.
plot(res_pp_iid)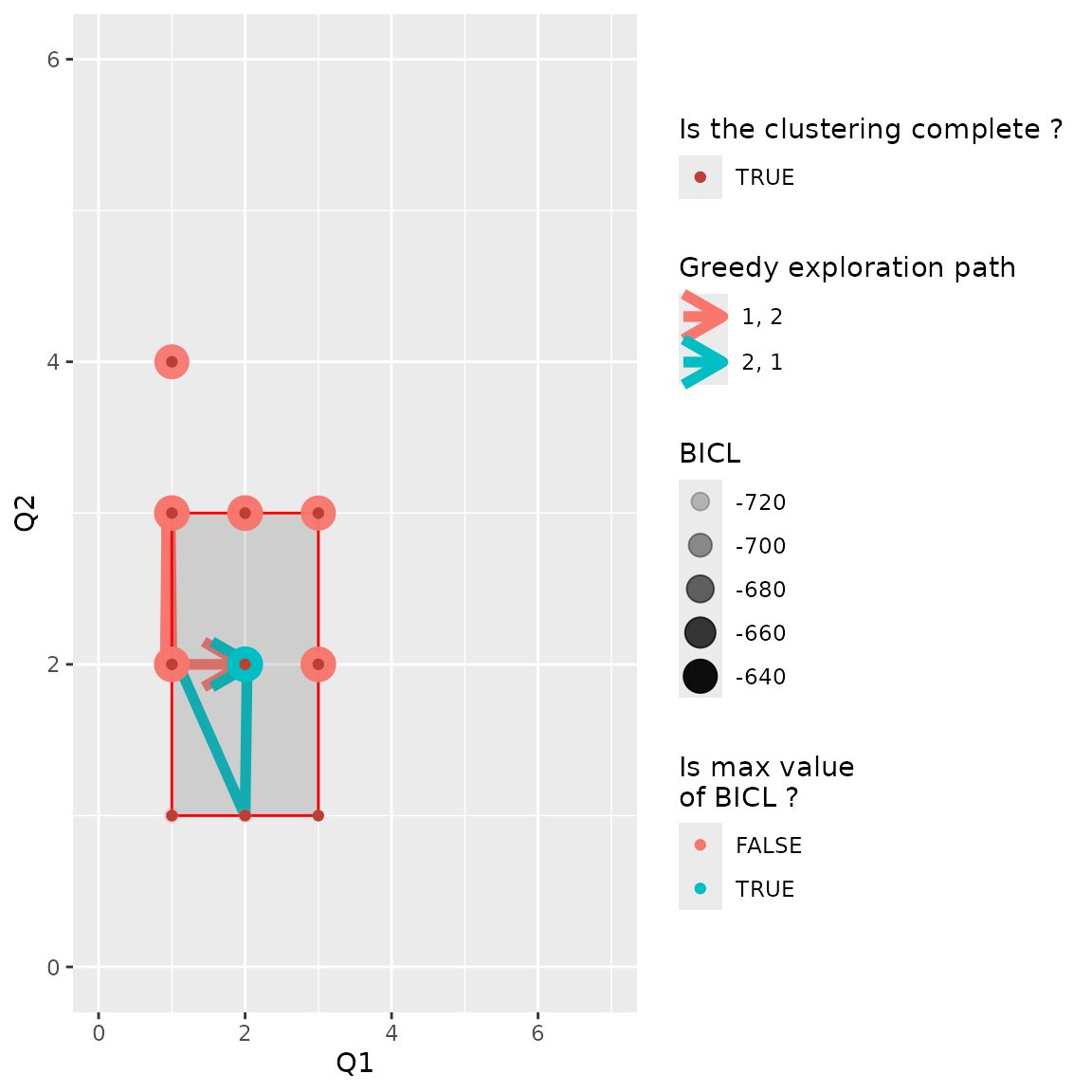
State-space exploration
best_fit <- res_pp_iid$best_fitResults and analysis
Here are some useful fields to analyze the results.
best_fit
#> Fitted Collection of Bipartite SBM -- bernoulli variant for 2 networks
#> =====================================================================
#> net_id = ( medan2002ld medan2002rb )
#> Dimensions = ( c(45, 21), c(72, 23) ) - ( 2, 2 ) blocks.
#> BICL = -633.8671
#> #Empty row blocks on all networks: 0 -- #Empty columns blocks on all networks: 0
#> * Useful fields
#> $distribution, $nb_nodes, $nb_blocks, $support, $prob_memberships
#> $memberships, $parameters, $BICL, $vbound, $pred_dyads
#> =====================================================================We can retrieve:
- the estimation of the model parameters
best_fit$parameters
#> $alpha
#> [,1] [,2]
#> [1,] 0.2499130 0.18198053
#> [2,] 0.4218242 0.03428153
#>
#> $pi
#> $pi[[1]]
#> [1] 0.06654147 0.93345853
#>
#> $pi[[2]]
#> [1] 0.06654147 0.93345853
#>
#>
#> $rho
#> $rho[[1]]
#> [1] 0.08387959 0.91612041
#>
#> $rho[[2]]
#> [1] 0.08387959 0.91612041- The block memberships:
best_fit$memberships[[2]]$row[1:10]
#> Birds Trochilidae Sappho sparganura
#> 2
#> Coleoptera Buprestidae Buprestidae sp.
#> 2
#> Coleoptera Cantharidae Cantharidae sp.
#> 2
#> Coleoptera Coccinellidae Coccinelidae sp1
#> 2
#> Coleoptera Coccinellidae Coccinelidae sp2
#> 2
#> Coleoptera Coccinellidae Coccinelidae sp3
#> 2
#> Coleoptera Curculionidae Curculionidae sp.
#> 2
#> Coleoptera Meloidae Epicauta sp.
#> 2
#> Coleoptera Mordellidae Mordellidae sp.
#> 2
#> Diptera Anthomyiidae Anthomyiidae sp2
#> 2And their probabilities:
best_fit$prob_memberships[[2]][[1]][1:10, 1]
#> Birds Trochilidae Sappho sparganura
#> 0.1369510749
#> Coleoptera Buprestidae Buprestidae sp.
#> 0.0117576955
#> Coleoptera Cantharidae Cantharidae sp.
#> 0.0019309149
#> Coleoptera Coccinellidae Coccinelidae sp1
#> 0.0008912515
#> Coleoptera Coccinellidae Coccinelidae sp2
#> 0.0019309149
#> Coleoptera Coccinellidae Coccinelidae sp3
#> 0.0019309149
#> Coleoptera Curculionidae Curculionidae sp.
#> 0.0019309149
#> Coleoptera Meloidae Epicauta sp.
#> 0.0251541553
#> Coleoptera Mordellidae Mordellidae sp.
#> 0.0008912515
#> Diptera Anthomyiidae Anthomyiidae sp2
#> 0.0019309149- The prediction for each dyads in the networks
best_fit$pred_dyads[[2]][1:10, 1]
#> Birds Trochilidae Sappho sparganura
#> 0.05504518
#> Coleoptera Buprestidae Buprestidae sp.
#> 0.03661664
#> Coleoptera Cantharidae Cantharidae sp.
#> 0.03517014
#> Coleoptera Coccinellidae Coccinelidae sp1
#> 0.03501710
#> Coleoptera Coccinellidae Coccinelidae sp2
#> 0.03517014
#> Coleoptera Coccinellidae Coccinelidae sp3
#> 0.03517014
#> Coleoptera Curculionidae Curculionidae sp.
#> 0.03517014
#> Coleoptera Meloidae Epicauta sp.
#> 0.03858861
#> Coleoptera Mordellidae Mordellidae sp.
#> 0.03501710
#> Diptera Anthomyiidae Anthomyiidae sp2
#> 0.03517014We can also plot the networks individually:
plot(res_pp_iid$best_fit, type = "block", net_id = 1) +
plot(res_pp_iid$best_fit, type = "block", net_id = 2)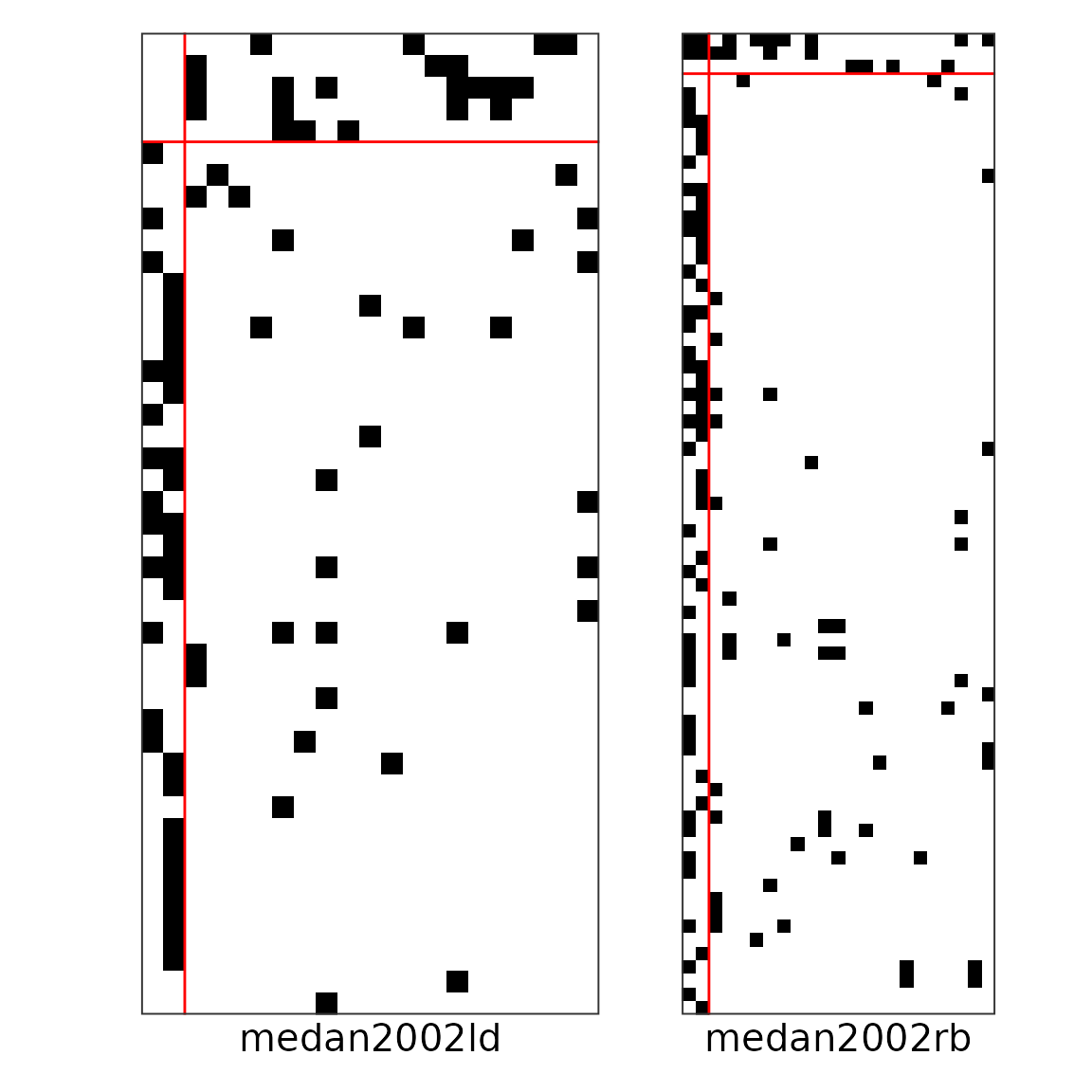
Networks after fitting the model and reordering the nodes and blocks
Or make different plots to exhibit the mesoscale structure:
plot(res_pp_iid$best_fit, type = "graphon")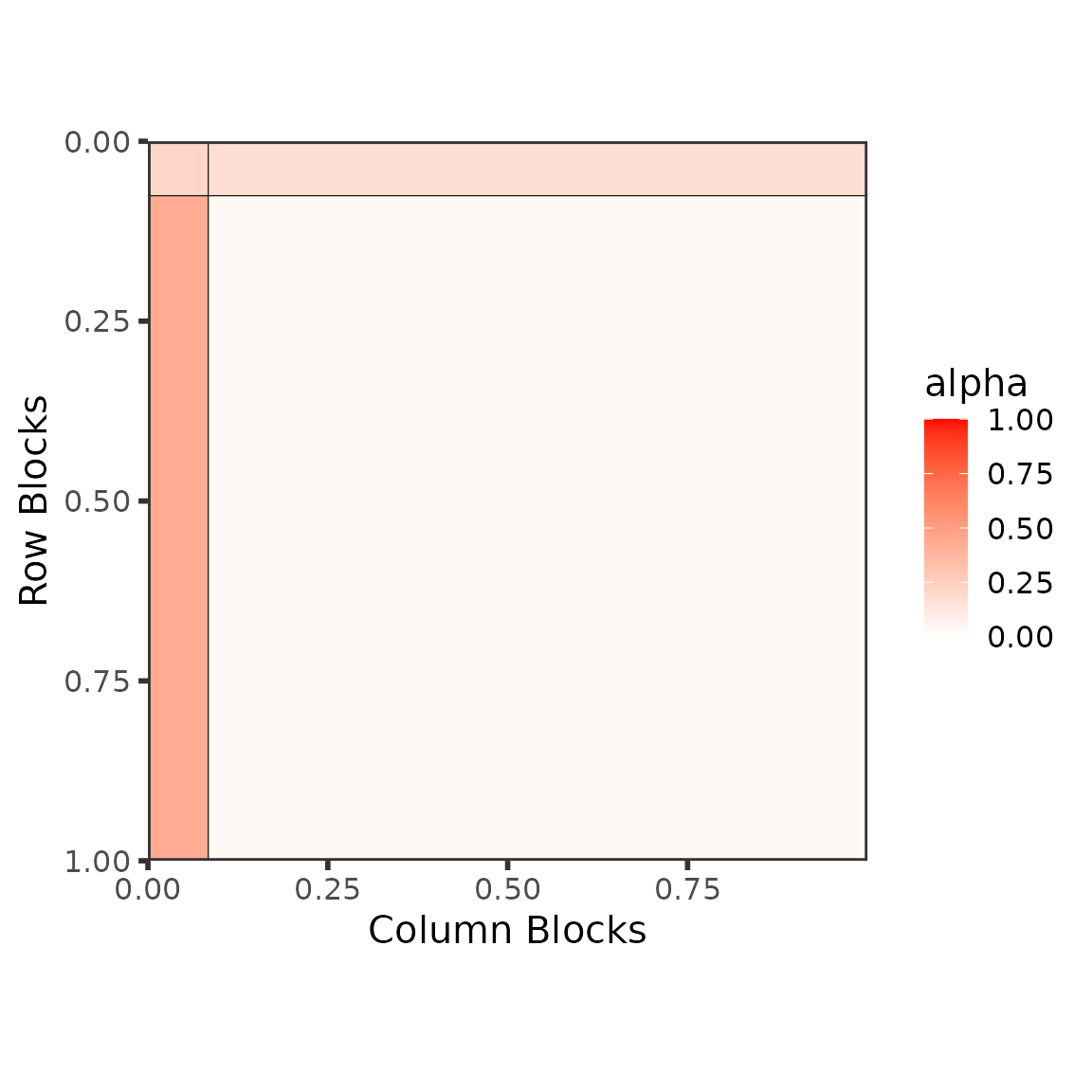
Graphon type plot
plot(res_pp_iid$best_fit, type = "meso", mixture = TRUE)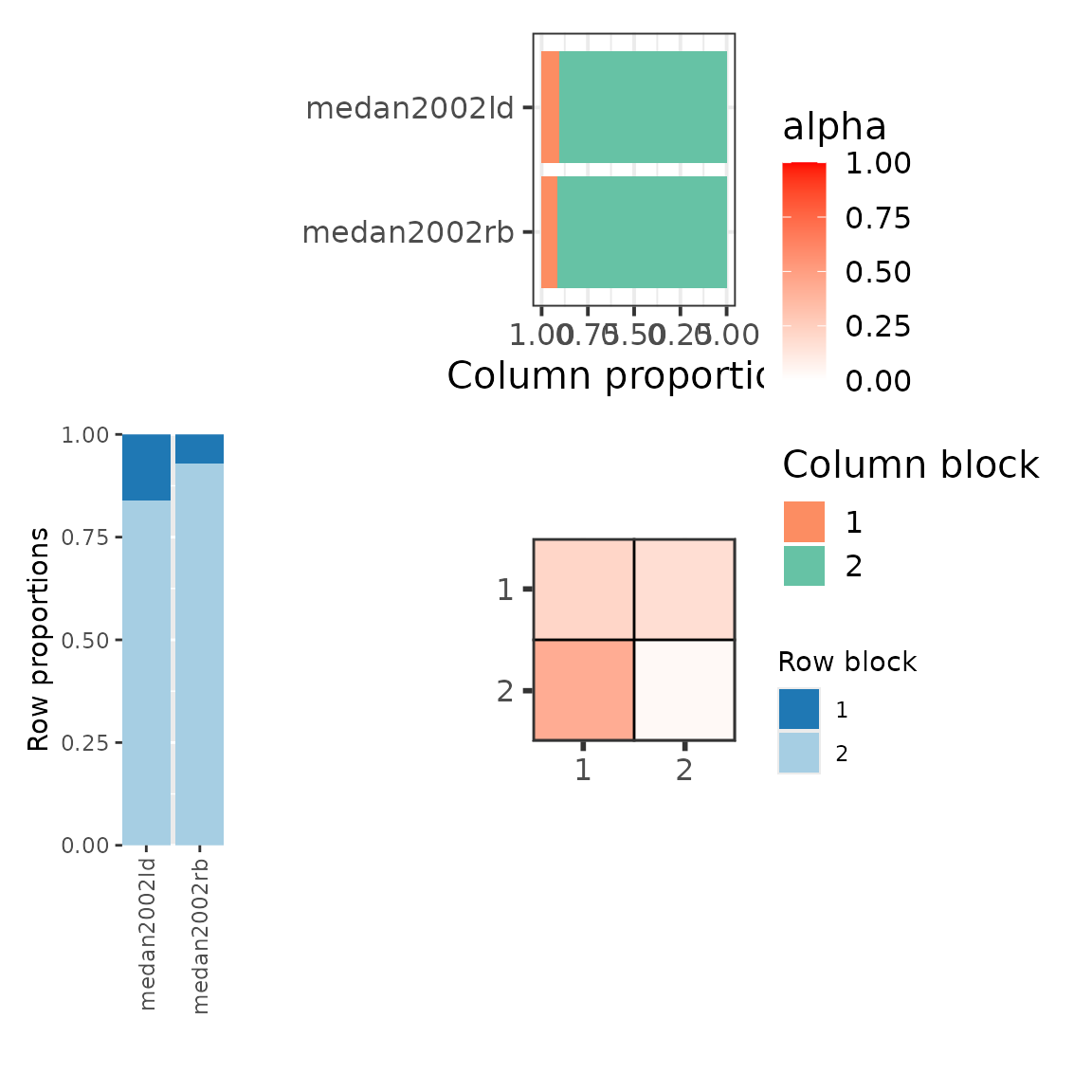
Mesoscale type plot
Networks not benefiting from joint modelisation
Next, we model jointly the medan2002ld, medan2002rb, olensen2002aig, olensen2002flo networks, using the iid-colBiSBM model.
res_pp_iid_sep <- estimate_colBiSBM(
netlist = dorebipartite[7L:10L], # A list of networks
colsbm_model = "iid", # The name of the model
net_id = names(dorebipartite)[7L:10L], # Name of the networks
nb_run = 1L, # Number of runs of the algorithm
global_opts = list(
verbosity = 1L,
plot_detail = 0L,
nb_cores = 2L,
backend = "no_mc"
)
)
#>
#>
#>
#>
#>
#> ==== Best fits criterion for the 4 networks. Computed in 1.03 secs ====
#> Sep BiSBM total BICL: -813.0666
#> colBiSBM BICL: -816.8325
#> Separated modelisation preferred.
#>
#> ==== Full computation performed in 20.9 secs ====The output indicates that the collection does not benefit from a joint modelisation.
Separated modelisation preferred
The structures might be too different to be gathered in one collection.
Clustering of networks
In the case of different structures clustering can be used to find a partitionning among all the networks.
We will simulate networks and add them to the 4 networks we used previously.
alpha <- matrix(c(
0.9, 0.55,
0.6, 0.1
), 2, 2, byrow = TRUE)
pi <- c(0.73, 0.27)
rho <- c(0.75, 0.25)
sim_net <-
generate_bipartite_collection(
nr = 40L,
nc = 30L,
pi = pi,
rho = rho,
alpha = alpha,
M = 2L,
model = "iid"
)
set.seed(1234L)
net_clust <- clusterize_bipartite_networks(
netlist = c(dorebipartite[7L:10L], sim_net), # A list of networks
colsbm_model = "iid", # The name of the model
net_id = c(
names(dorebipartite)[7L:10L],
paste0("sim", seq_along(sim_net))
), # Name of the networks
nb_run = 1L, # Number of runs of the algorithm
global_opts = list(
verbosity = 0L,
plot_details = 0L,
nb_cores = 2L,
backend = "no_mc",
Q1_max = 9L,
Q2_max = 9L
)
)
#> ℹ A save file will be created at "/tmp/Rtmpaobtw0/file6a2d700c3191.Rds" and updated after each step
#>
#> ── Fitting the full collection ─────────────────────────────────────────────────
#>
#> ── Beginning clustering ────────────────────────────────────────────────────────
#>
#> ── Trying to split the collection of "medan2002ld", "medan2002rb", "olensen2002aig", "olensen2002flo", "sim1", and "sim2" ──
#>
#> ℹ Fitting a sub collection with : "medan2002ld", "medan2002rb", "olensen2002aig", and "olensen2002flo"
#> ℹ Fitting a sub collection with : "sim1" and "sim2"
#> ✔ Splitting collections improved the BIC-L criterion
#>
#> ── Trying to split the collection of "medan2002ld", "medan2002rb", "olensen2002aig", and "olensen2002flo" ──
#>
#> ℹ Fitting a sub collection with : "medan2002ld" and "medan2002rb"
#> ℹ Fitting a sub collection with : "olensen2002aig" and "olensen2002flo"
#>
#> The window is (partially) out of domain !
#> Trying to go from (1, 0) to (3, 2).
#> Max window should be :
#> (1, 9)---(9, 9)
#> || ||
#> (1, 1)---(9, 1)
#> The window will work best on valid configurations
#> ✔ Splitting collections improved the BIC-L criterion
#>
#> ── Trying to split the collection of "sim1" and "sim2" ──
#>
#> ℹ Fitting a sub collection with : "sim1"
#> ℹ Fitting a sub collection with : "sim2"
#> ✖ Splitting collections decreased the BIC-L criterion
#>
#> ── Trying to split the collection of "medan2002ld" and "medan2002rb" ──
#>
#> ℹ Fitting a sub collection with : "medan2002ld"
#> ℹ Fitting a sub collection with : "medan2002rb"
#>
#> The window is (partially) out of domain !
#> Trying to go from (0, 1) to (2, 3).
#> Max window should be :
#> (1, 9)---(9, 9)
#> || ||
#> (1, 1)---(9, 1)
#> The window will work best on valid configurations
#> ✖ Splitting collections decreased the BIC-L criterion
#>
#> ── Trying to split the collection of "olensen2002aig" and "olensen2002flo" ──
#>
#> ℹ Fitting a sub collection with : "olensen2002aig"
#>
#> The window is (partially) out of domain !
#> Trying to go from (1, 0) to (3, 2).
#> Max window should be :
#> (1, 9)---(9, 9)
#> || ||
#> (1, 1)---(9, 1)
#> The window will work best on valid configurations
#> ℹ Fitting a sub collection with : "olensen2002flo"
#>
#> The window is (partially) out of domain !
#> Trying to go from (0, 0) to (2, 2).
#> Max window should be :
#> (1, 9)---(9, 9)
#> || ||
#> (1, 1)---(9, 1)
#> The window will work best on valid configurations
#> ✖ Splitting collections decreased the BIC-L criterion
#> ✔ Finished clustering
#> ℹ After clustering the partition has a BIC-L of -2041.69410950077
#> ℹ The final results are saved at "/tmp/Rtmpaobtw0/file6a2d700c3191.Rds"
best_partition <- net_clust$partitionThe obtained partition below reveals that our 2 simulated networks are considered as being part of the same collection and that the among the 4 plant-pollinator networks there exists a difference leading to 2 collections that contains networks from the same authors.
The plot of the mesoscale structures of the 3 groups shows:
wrap_plots(
lapply(best_partition, function(collection) {
plot(collection, type = "graphon") +
ggplot2::ggtitle(label = "", subtitle = toString(collection$net_id))
}),
ncol = 2L
)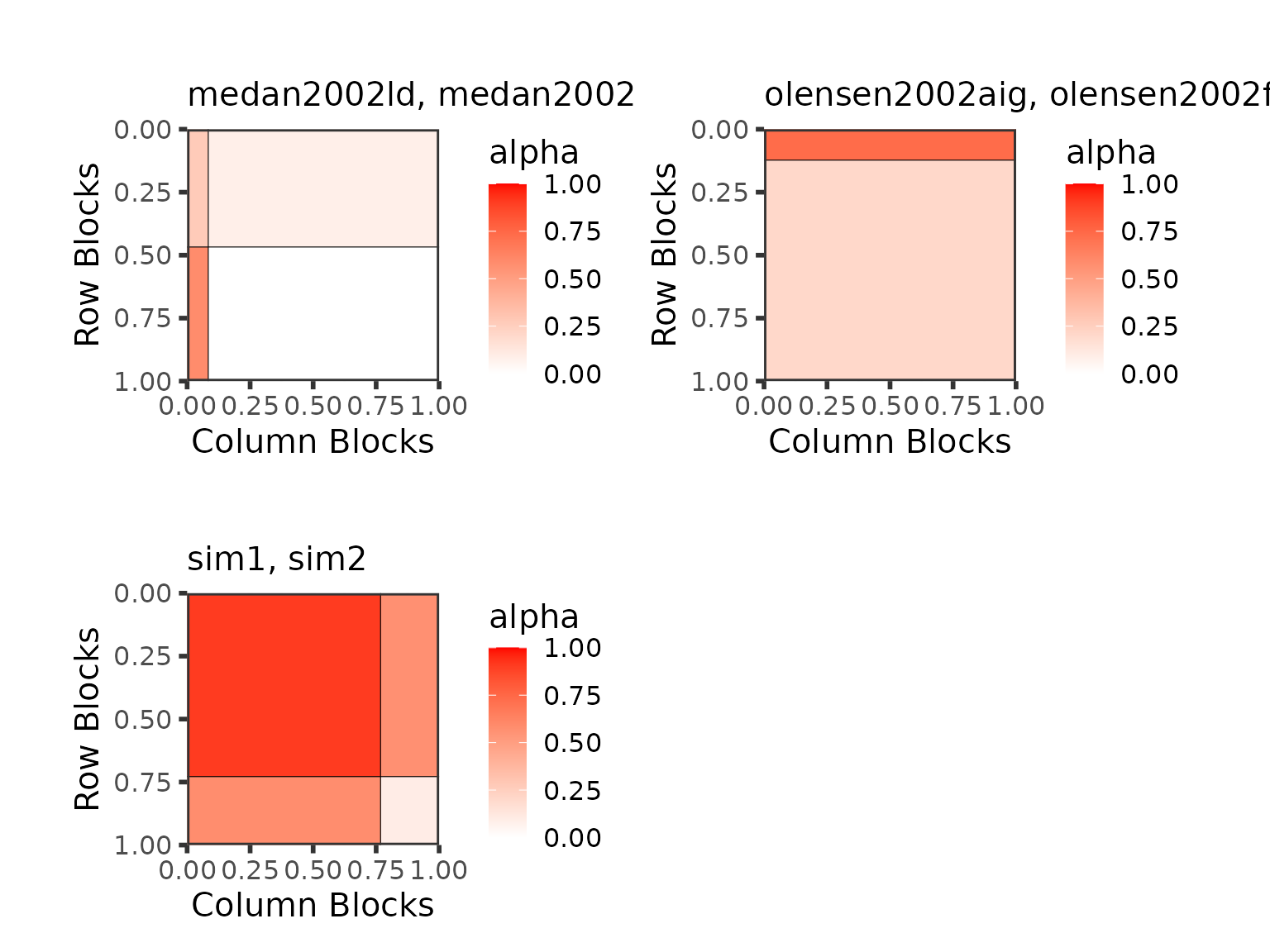
Best partition graphon type plots